Application Examples
Find example applications for your Verity Software House apps to get started quickly! Each example can be downloaded for your own use. Your are welcome to modify the examples to suit your lab's needs.
GemStone
GemStone Project for Cytek cFluor 14-C Immune Profiling Kit
GemStone Project for Cytek 33-C Immune Profiling
WinList
WinList Bundle for Cytek cFluor 14-C Immune Profiling Kit
WinList Bundle for Fluidigm Maxpar Direct Immune Profiling Kit
WinList Bundle shown in Merging Files and Cen-se Video
ModFit LT
GemStone Application Examples
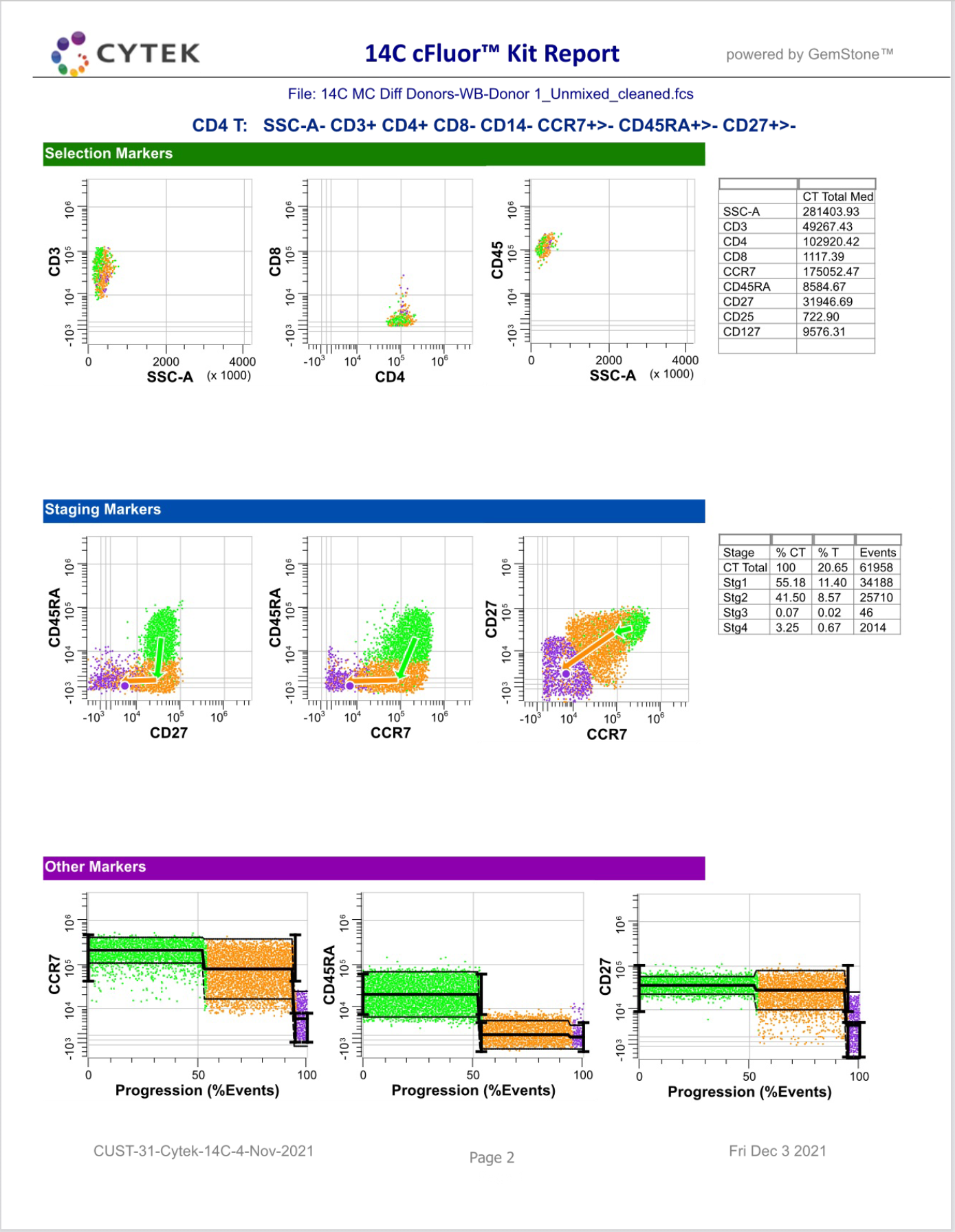
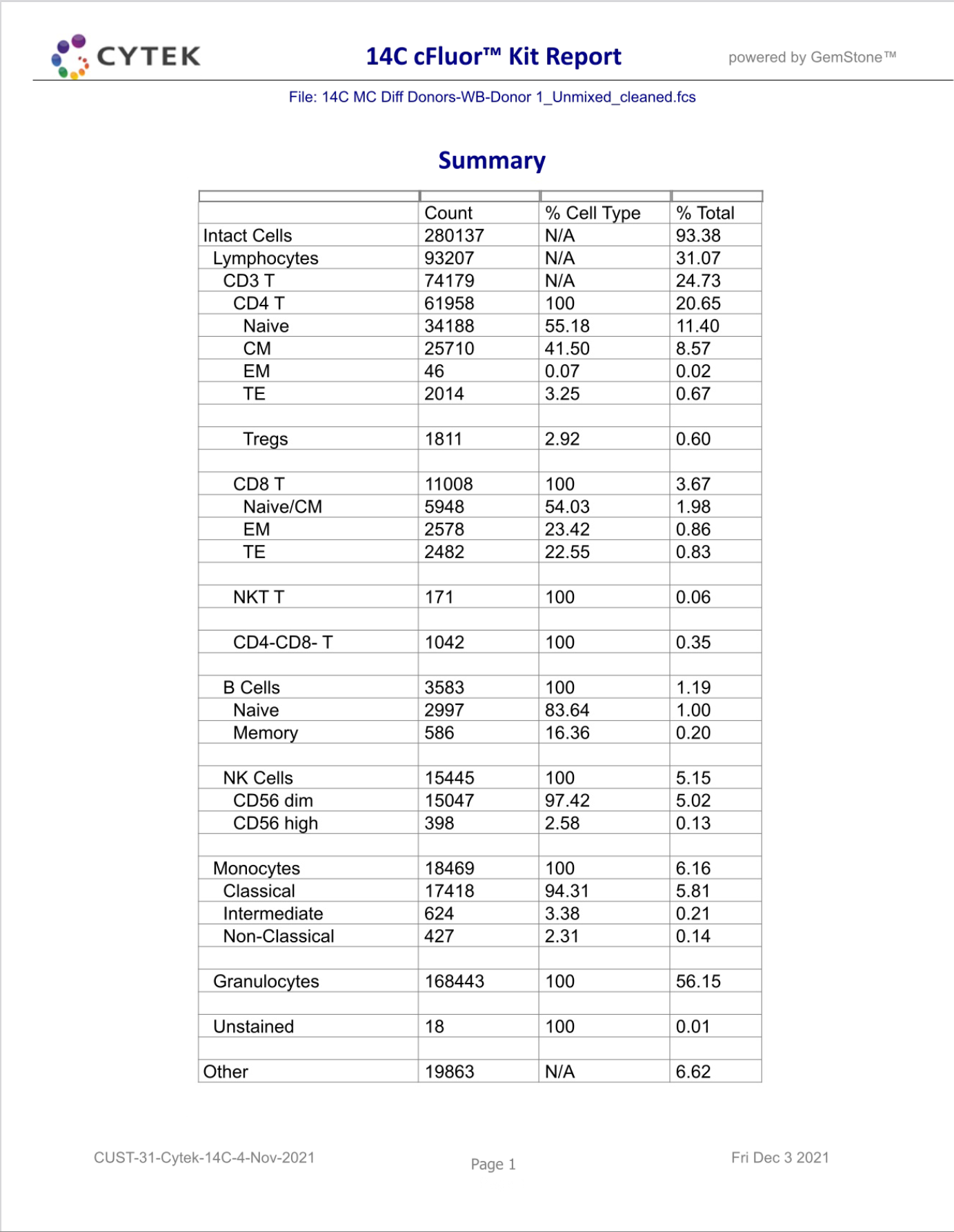
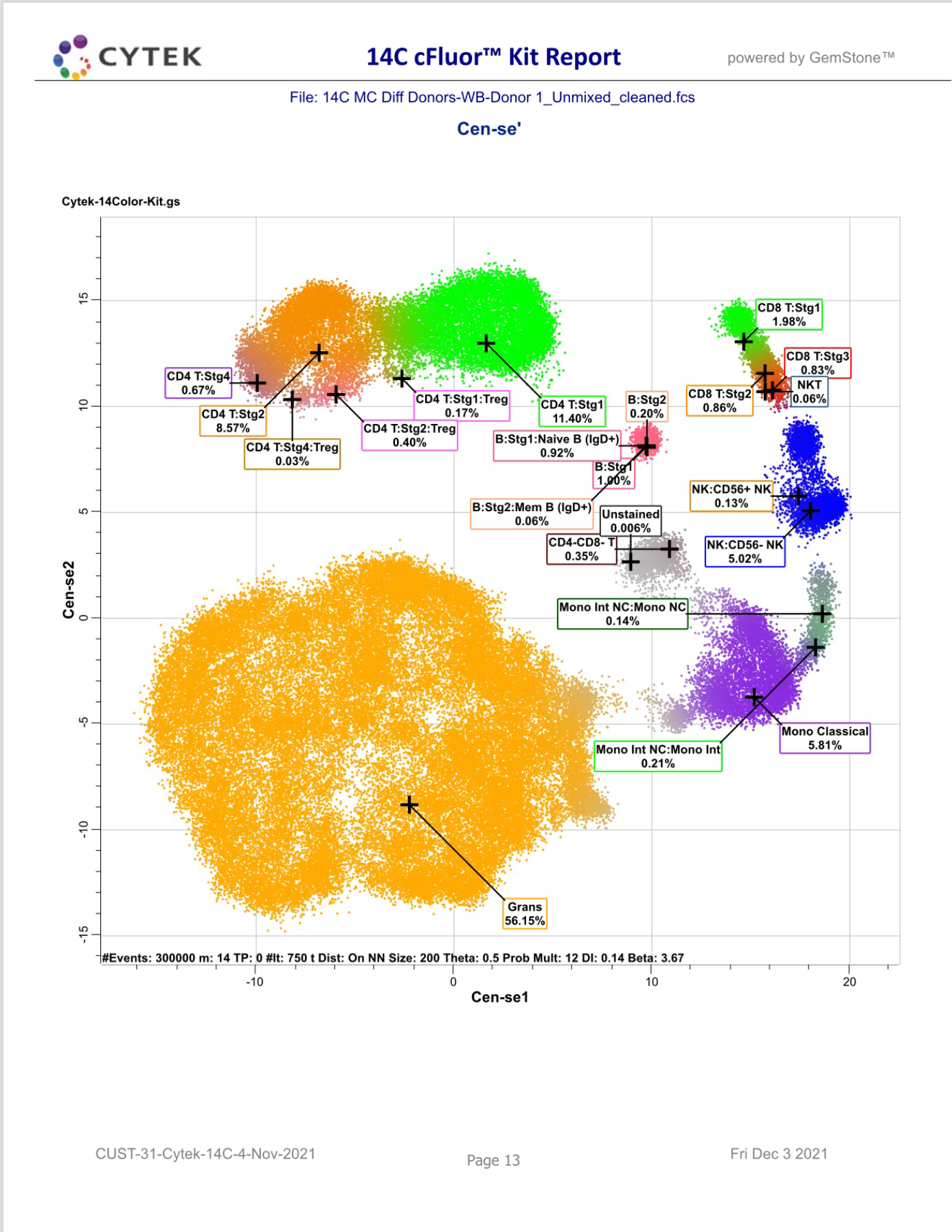
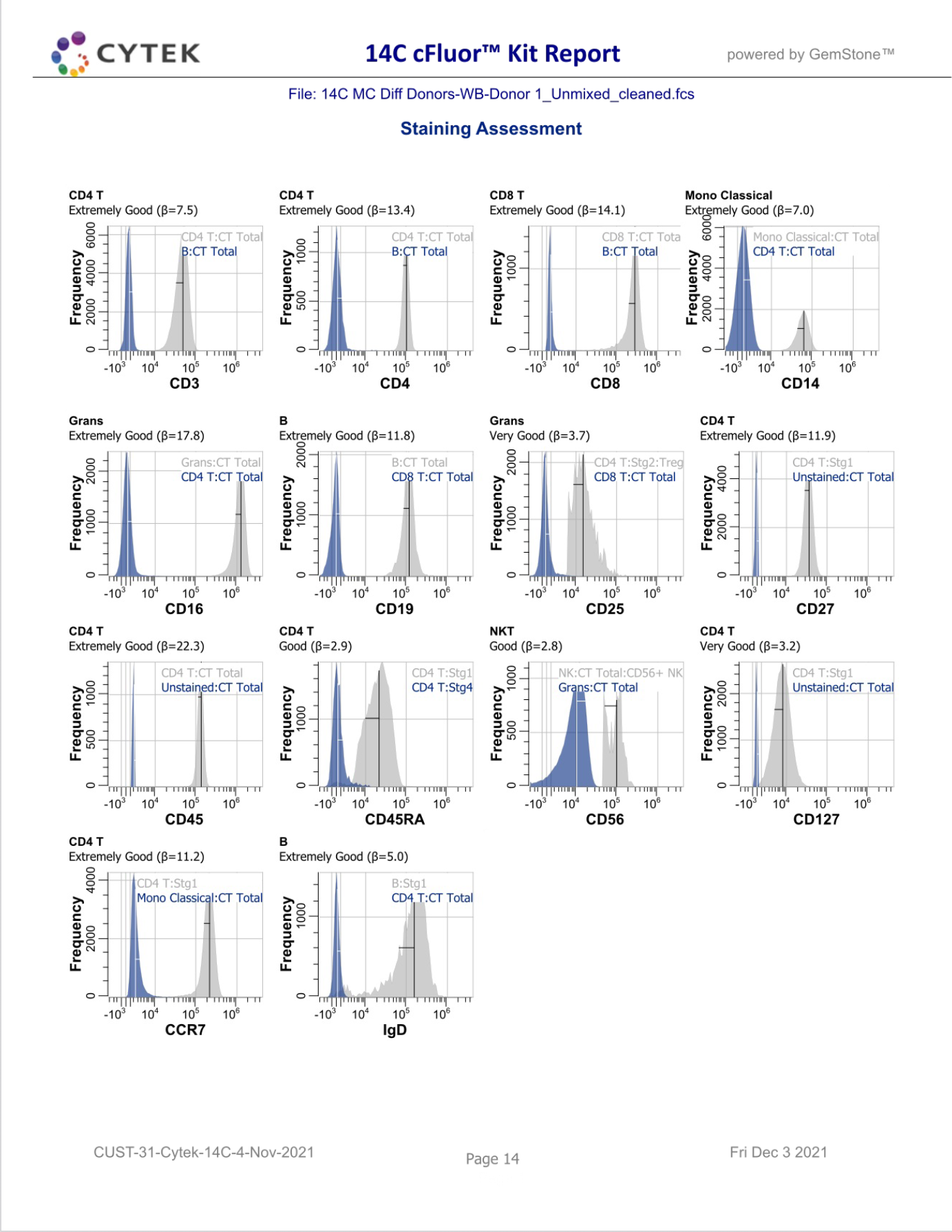
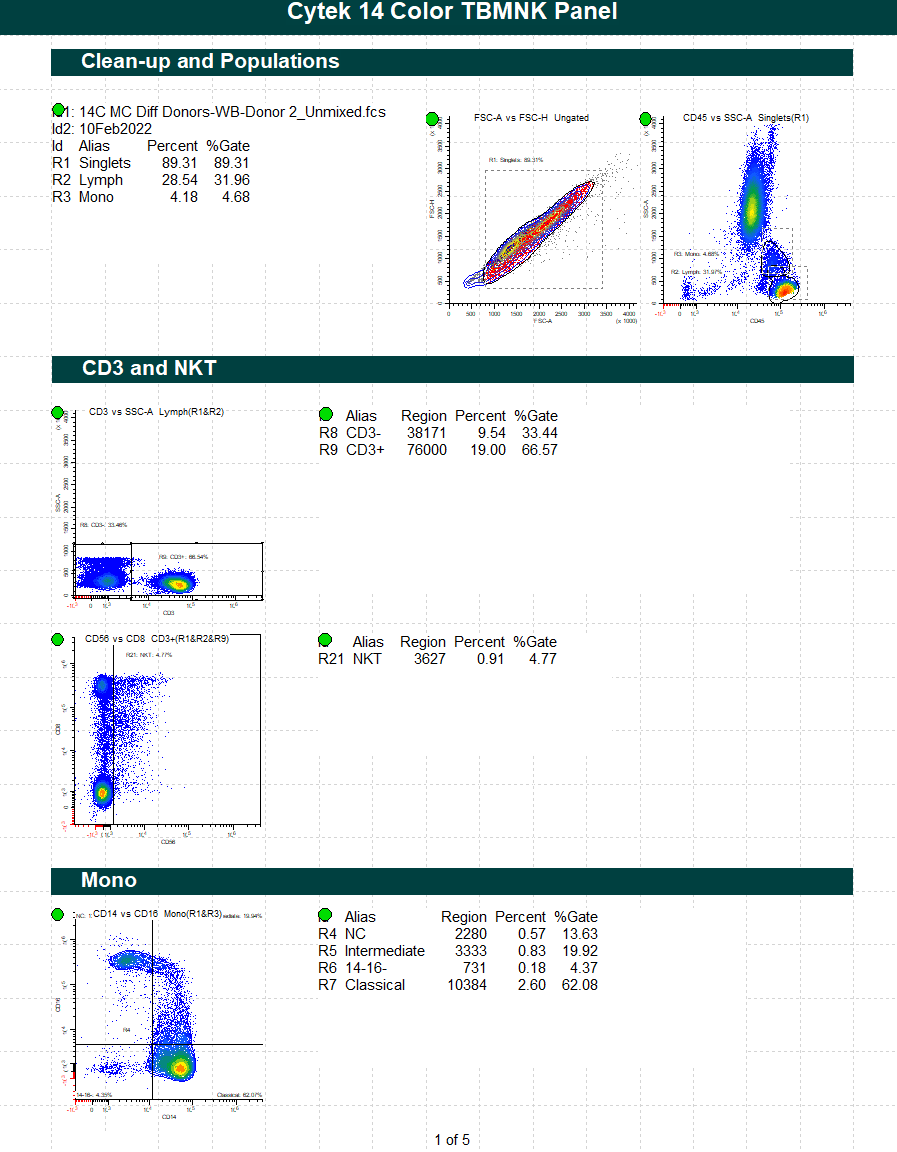
GemStone Project for Cytek cFluor 14-C Immune Profiling Kit
Use this project to analyze 14-color data from Cytek Aurora and the cFluor 14C kit (Cytek # R7-40000). The analysis quantifies CD4 T, CD8T, NK, NKT, B, monocytes, and granulocytes, with report pages for each. A Cen-se' dimensionality reduction plot is included, along with a staining assessment for each marker.
PDF reports are generated for clean-up and immunophenotyping results, and a database of all reported results.
For GemStone 2.0+

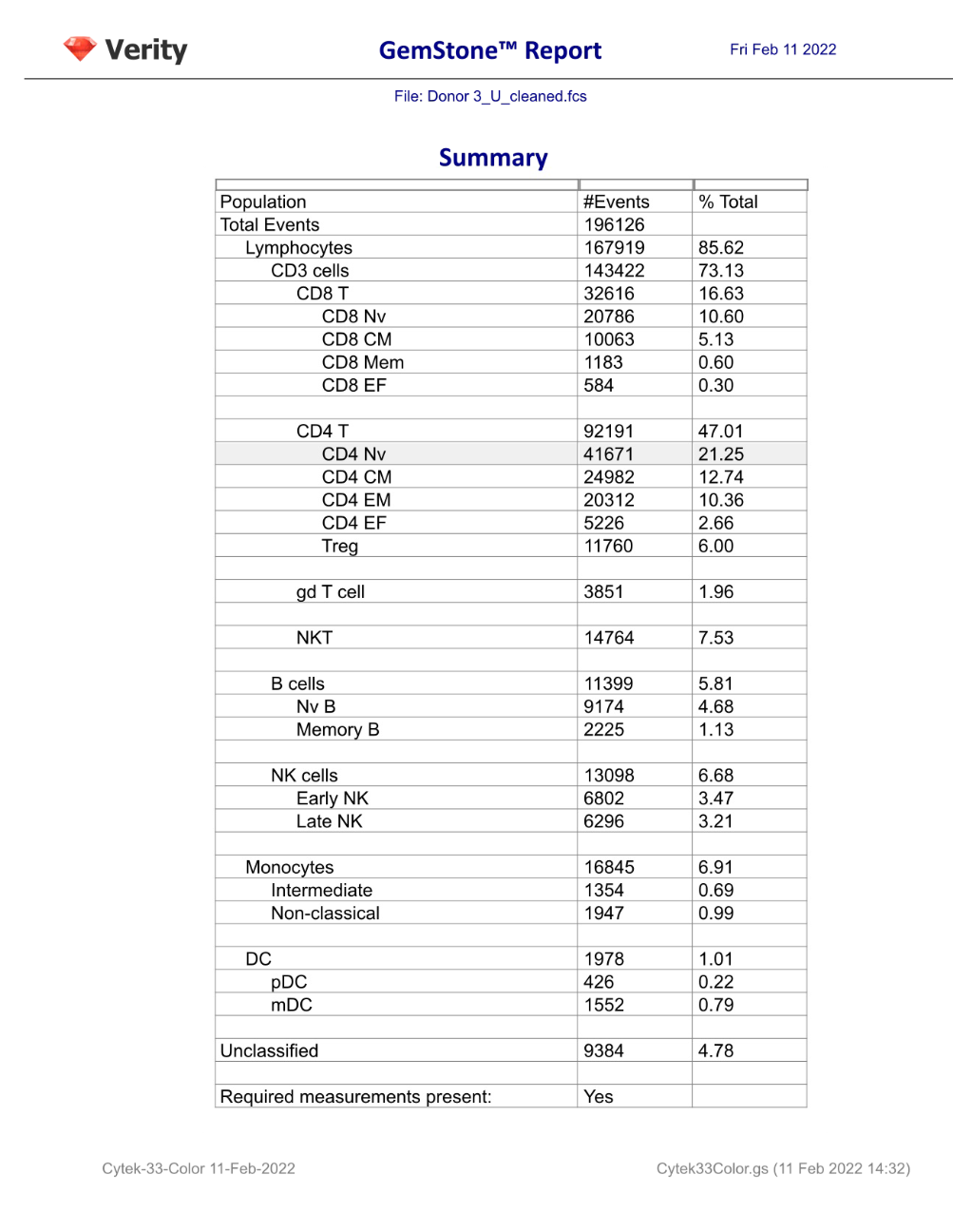
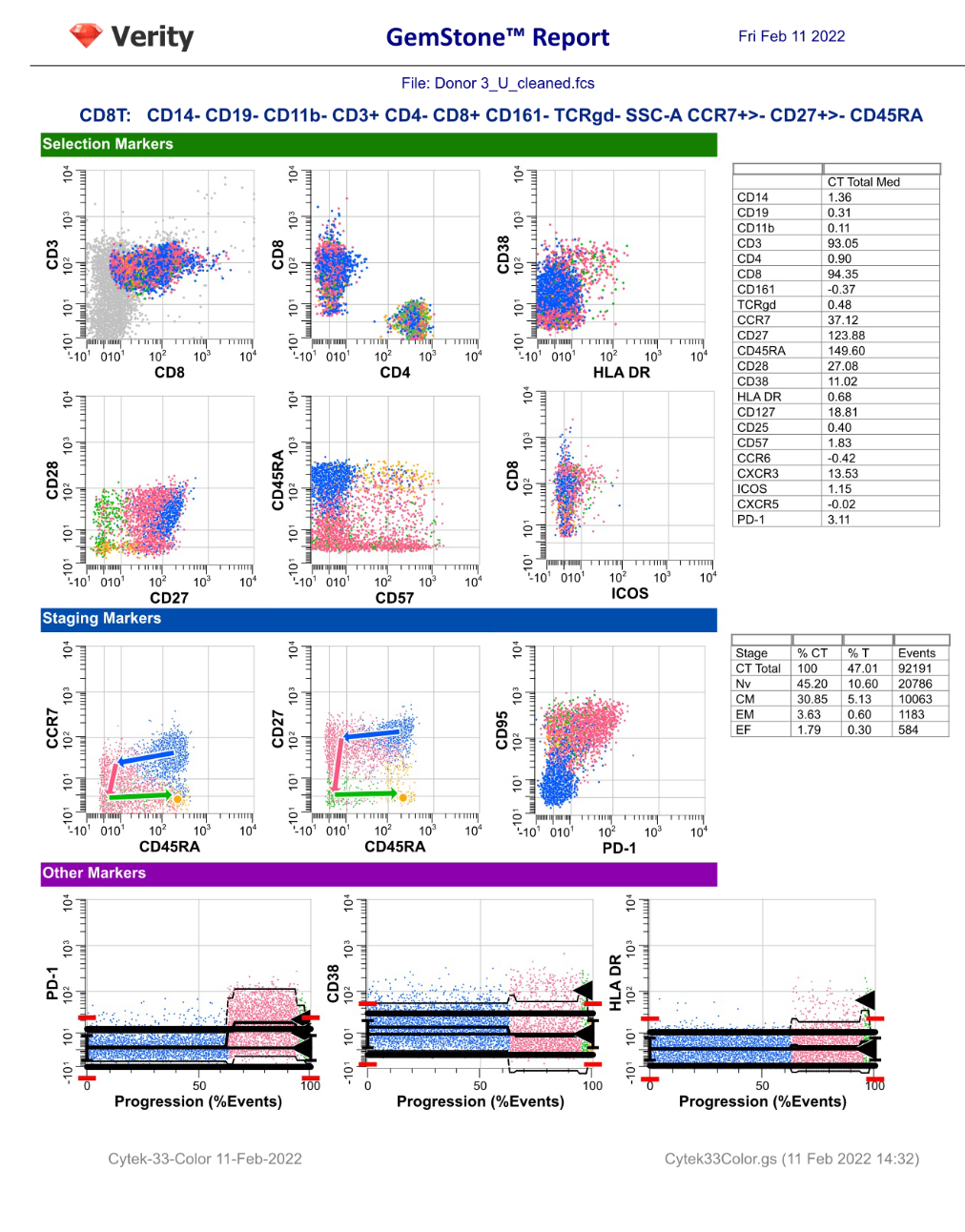
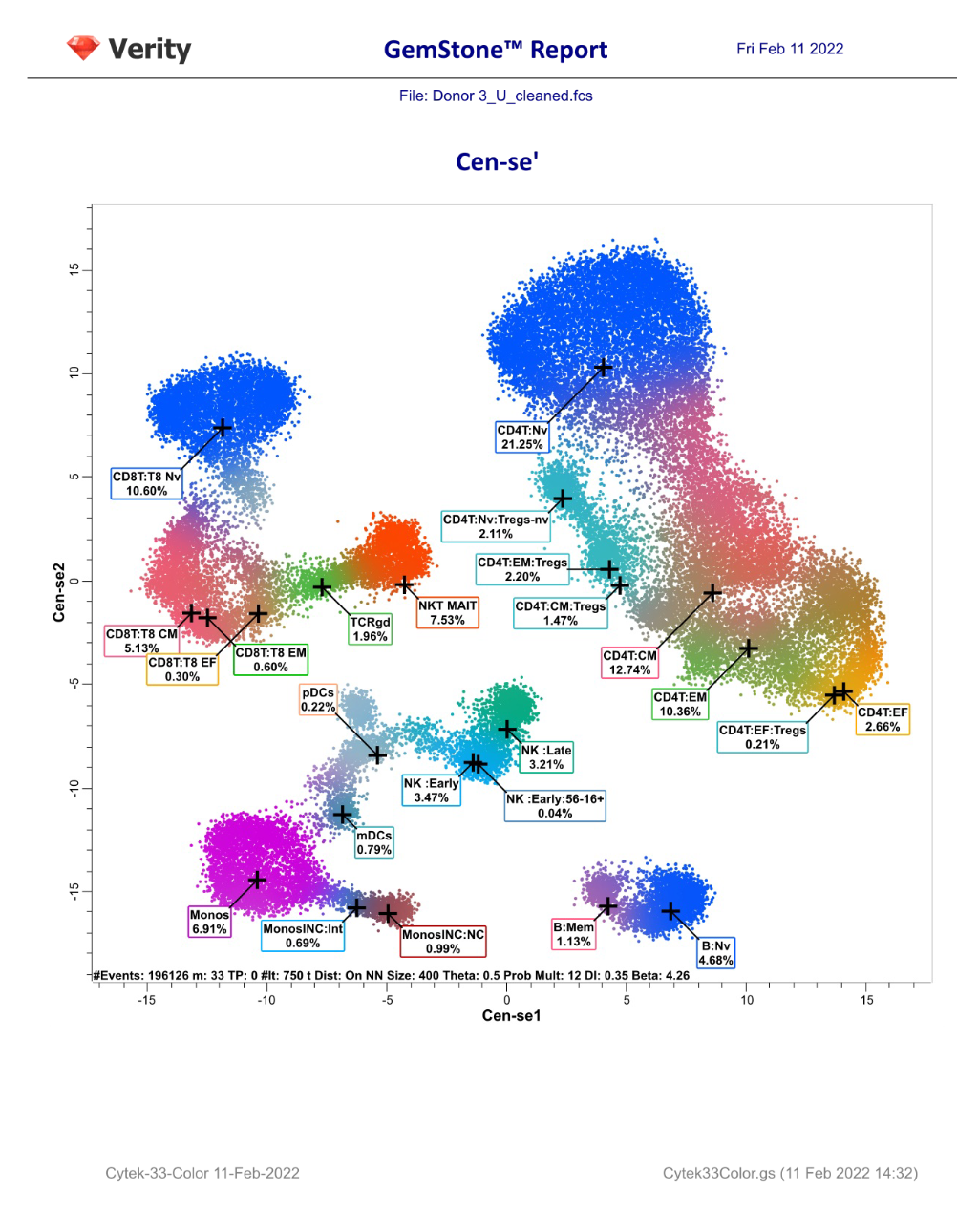
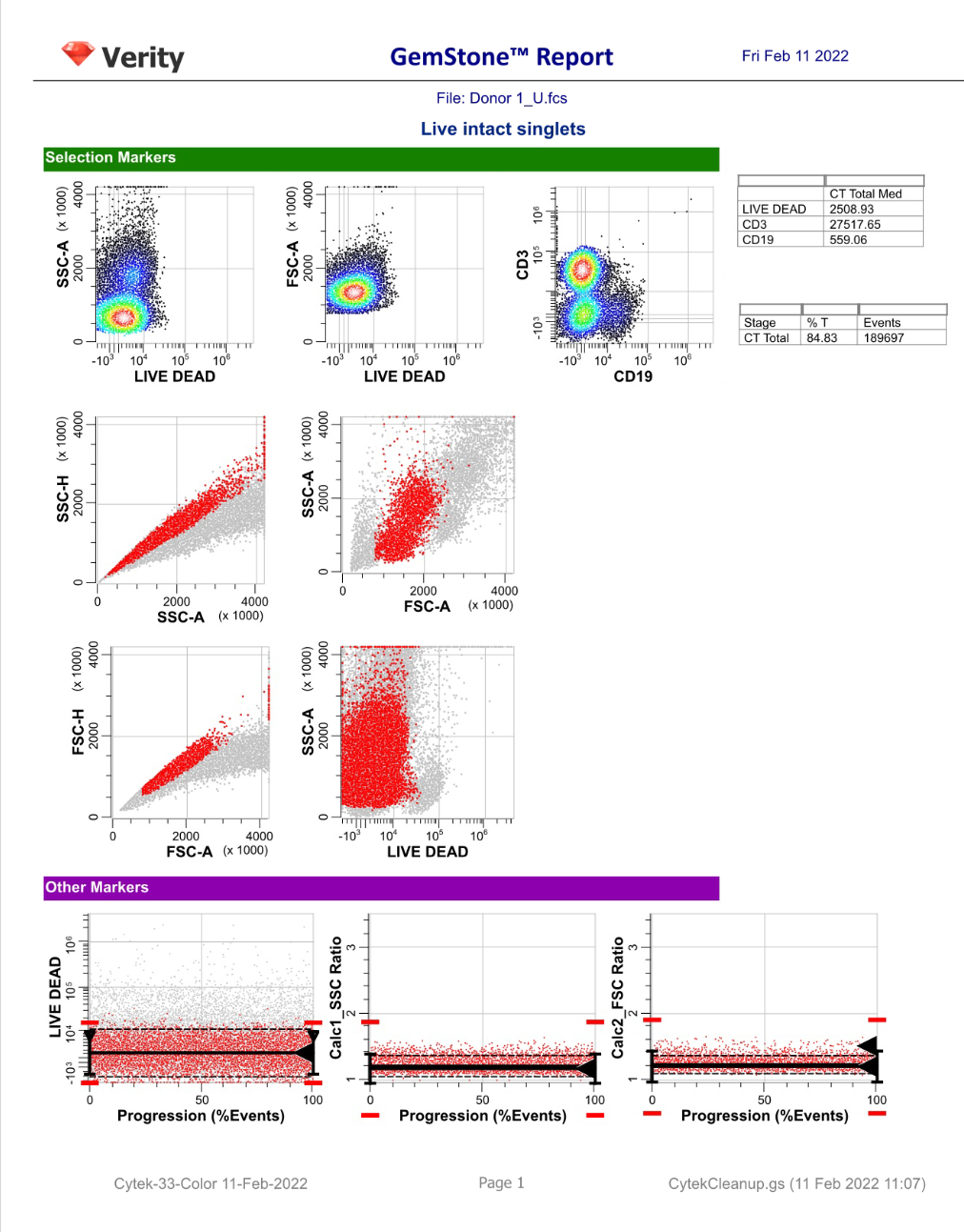
GemStone Project for Cytek 33-Color Immune Profiling
Use this project to analyze 33-color data from Cytek Aurora. Nine cell types are identified: CD4 T, CD8T, NKT/MAIT, gd T, B, NK, monocytes, pDCs, and mDCs. The project is two-pass: the first to clean-up, the second to analyze the cell types. A Cen-se' dimensionality reduction plot is included, along with a staining assessment for each marker.
Output includes PDF reports for clean-up and immunophenotyping, and a database of all reported results.
For GemStone 2.0+

WinList Application Examples
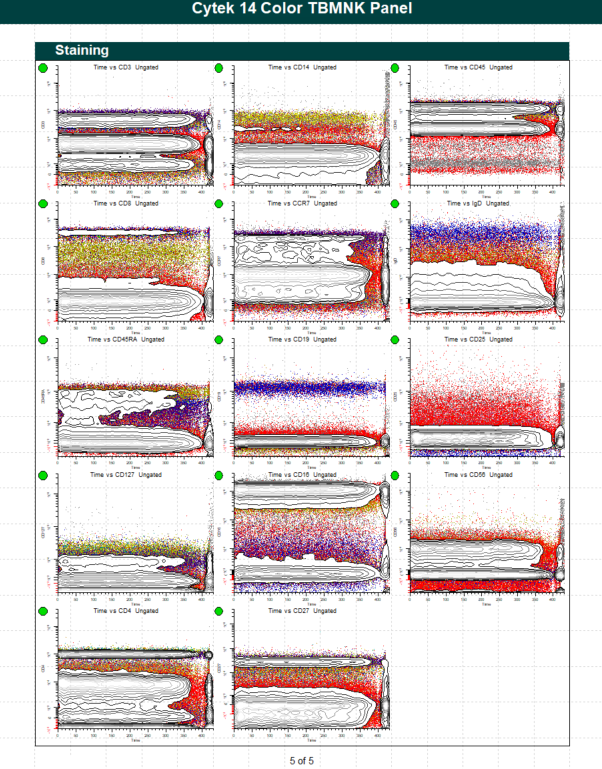
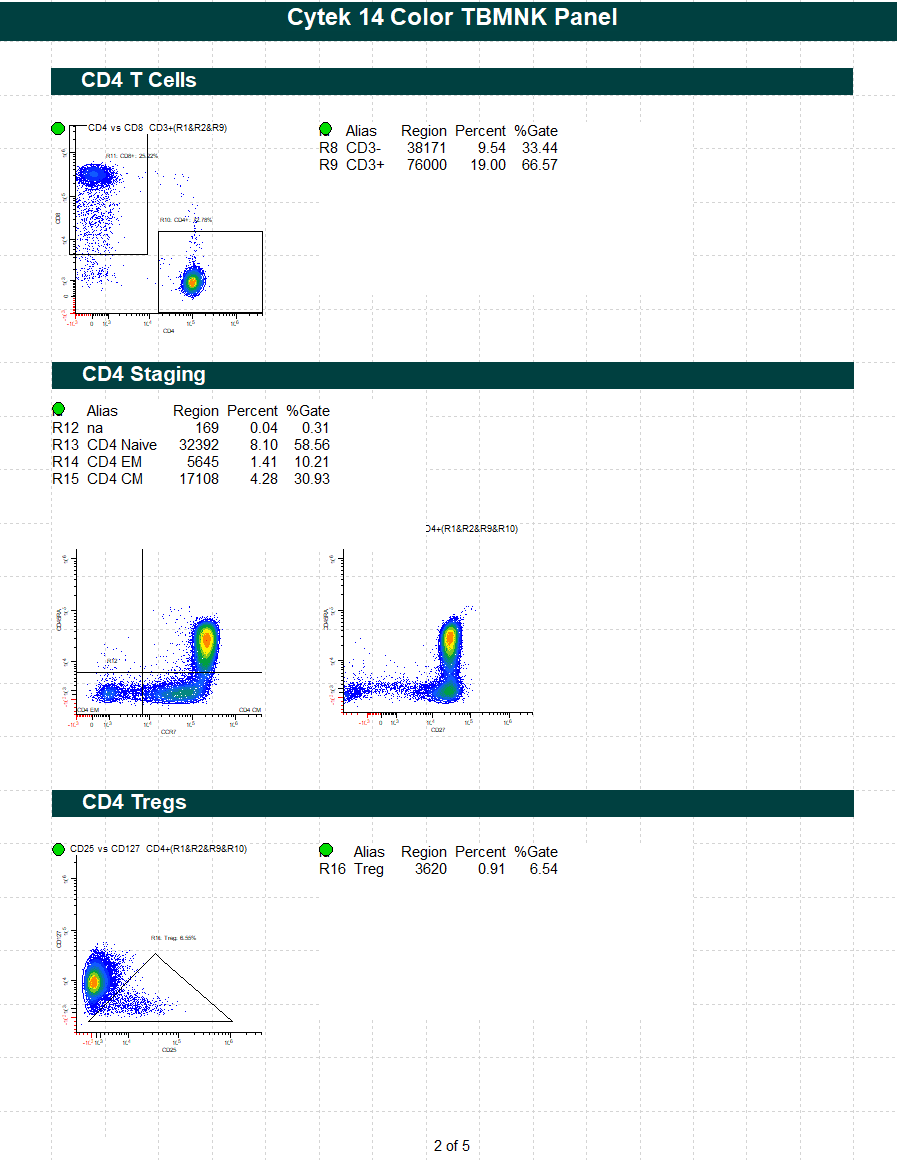
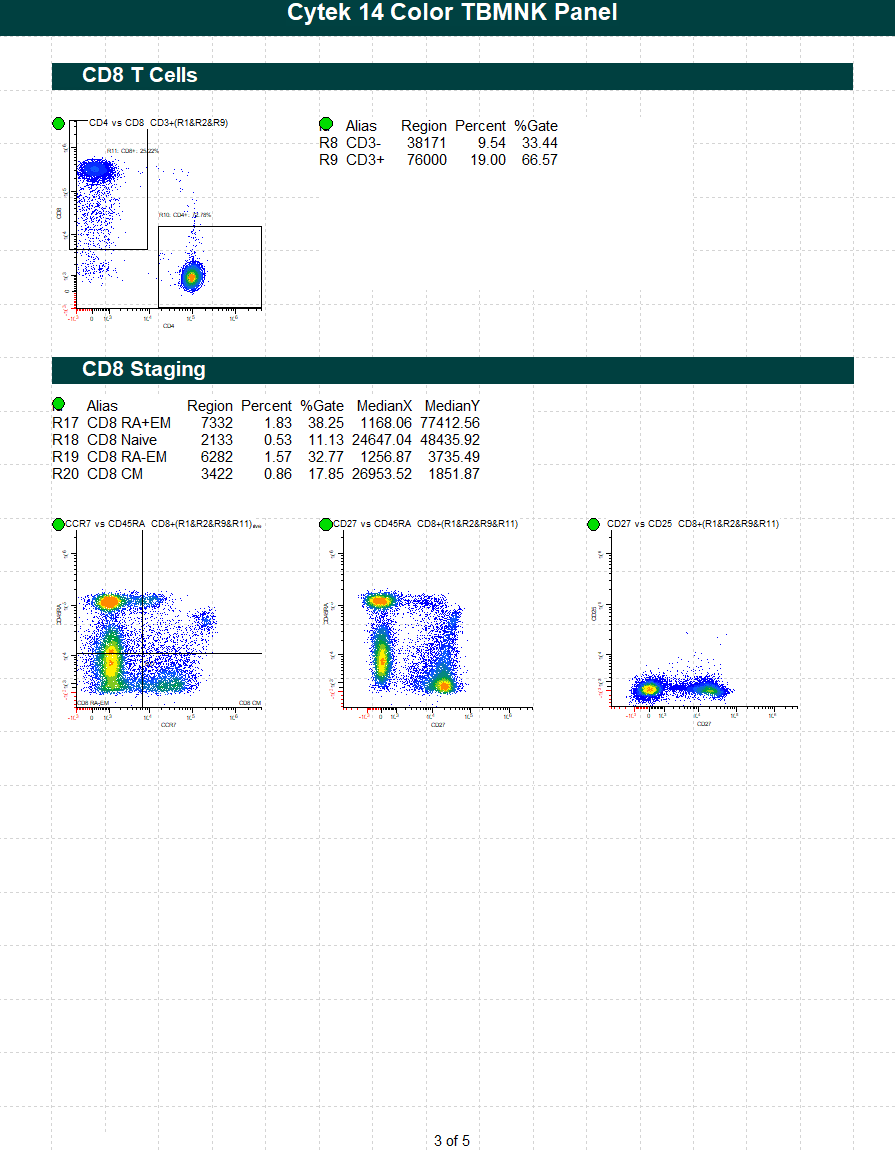
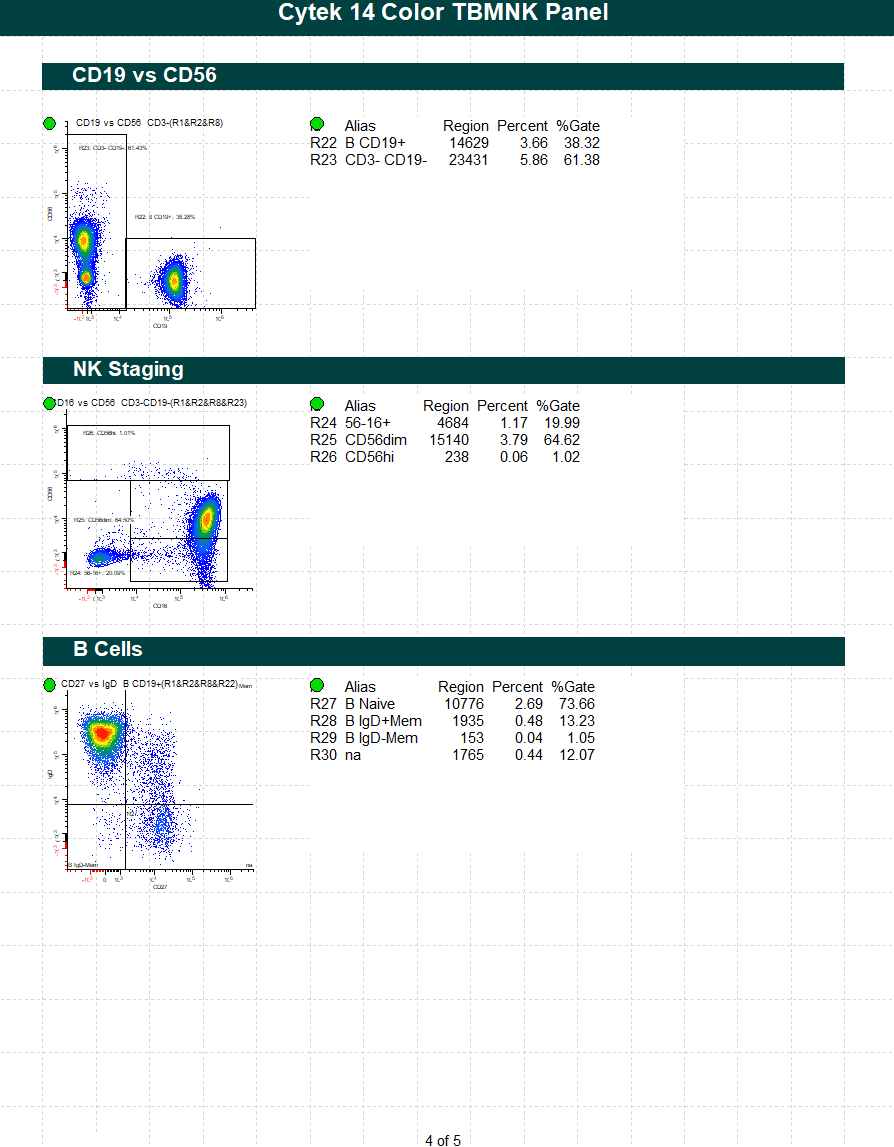
WinList Bundle for Cytek cFluor 14-C Immune Profiling Kit
This protocol bundle analyzes 14-color data from Cytek Aurora and the cFluor 14C kit (Cytek # R7-40000). The analysis quantifies CD4 T, CD8T, NK, NKT, B, and monocytes, and granulocytes. A stability assessment for each marker is included.
The protocol makes use of probability contour regions to automatically identify singlets, monos, and lymphs.
For WinList 10+
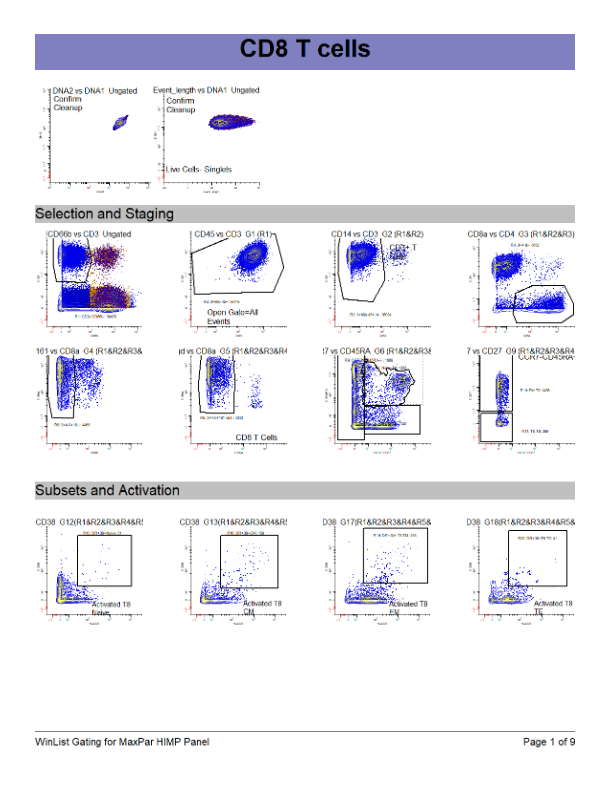
WinList Bundle for Fluidigm Maxpar Direct Immune Profiling Kit
Use this protocol bundle to analyze data from Fluidigm's Maxpar Direct Immune Profiling Assay (Fluidigm # 201325). The analysis protocol is designed for 30 antibodies and identifies 37 immune cell populations.
This is a handy tool for comparison of gating with other analytical approaches, such as Maxpar Pathsetter. A nine page report is included.
For WinList 10+

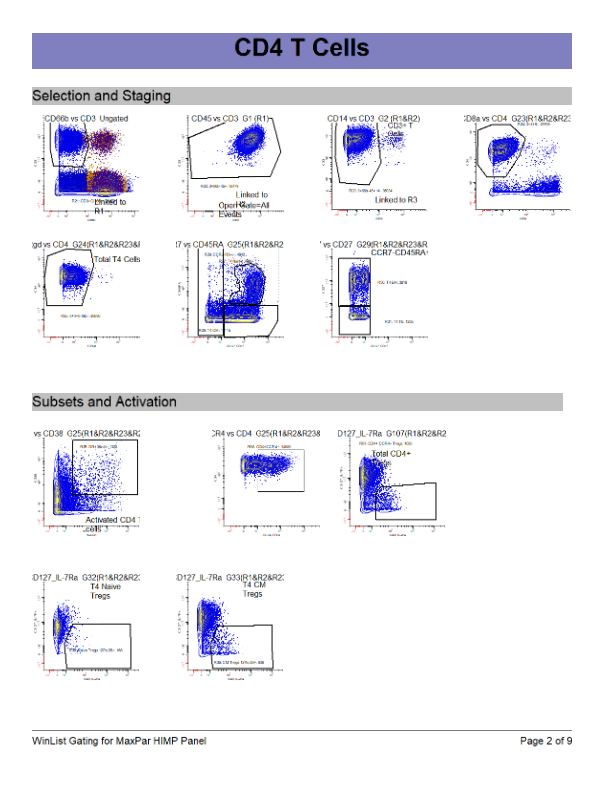
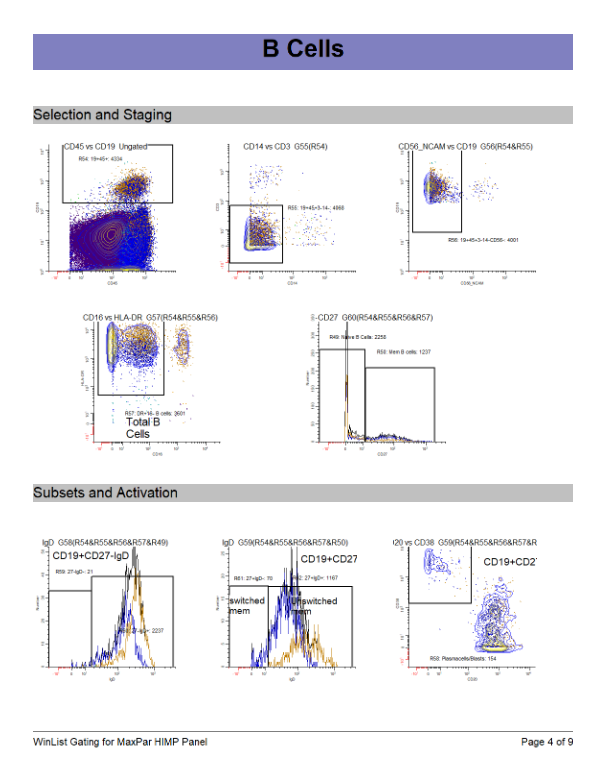
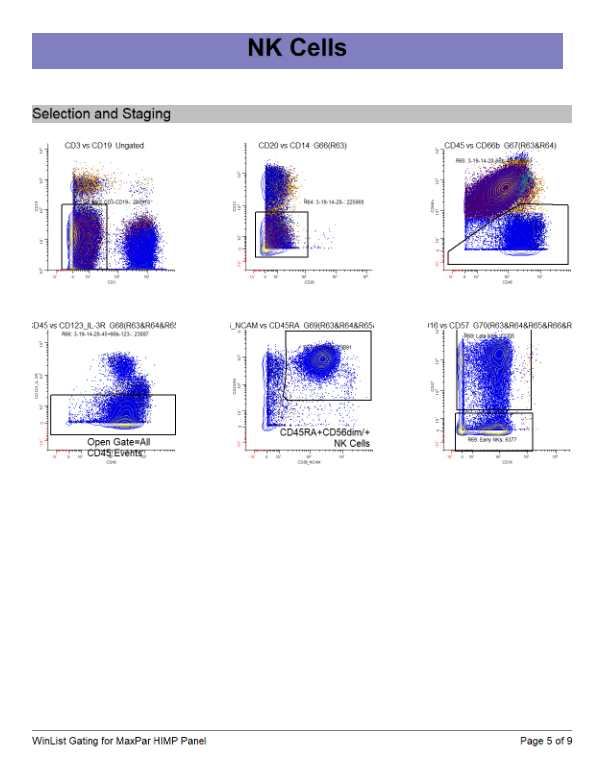
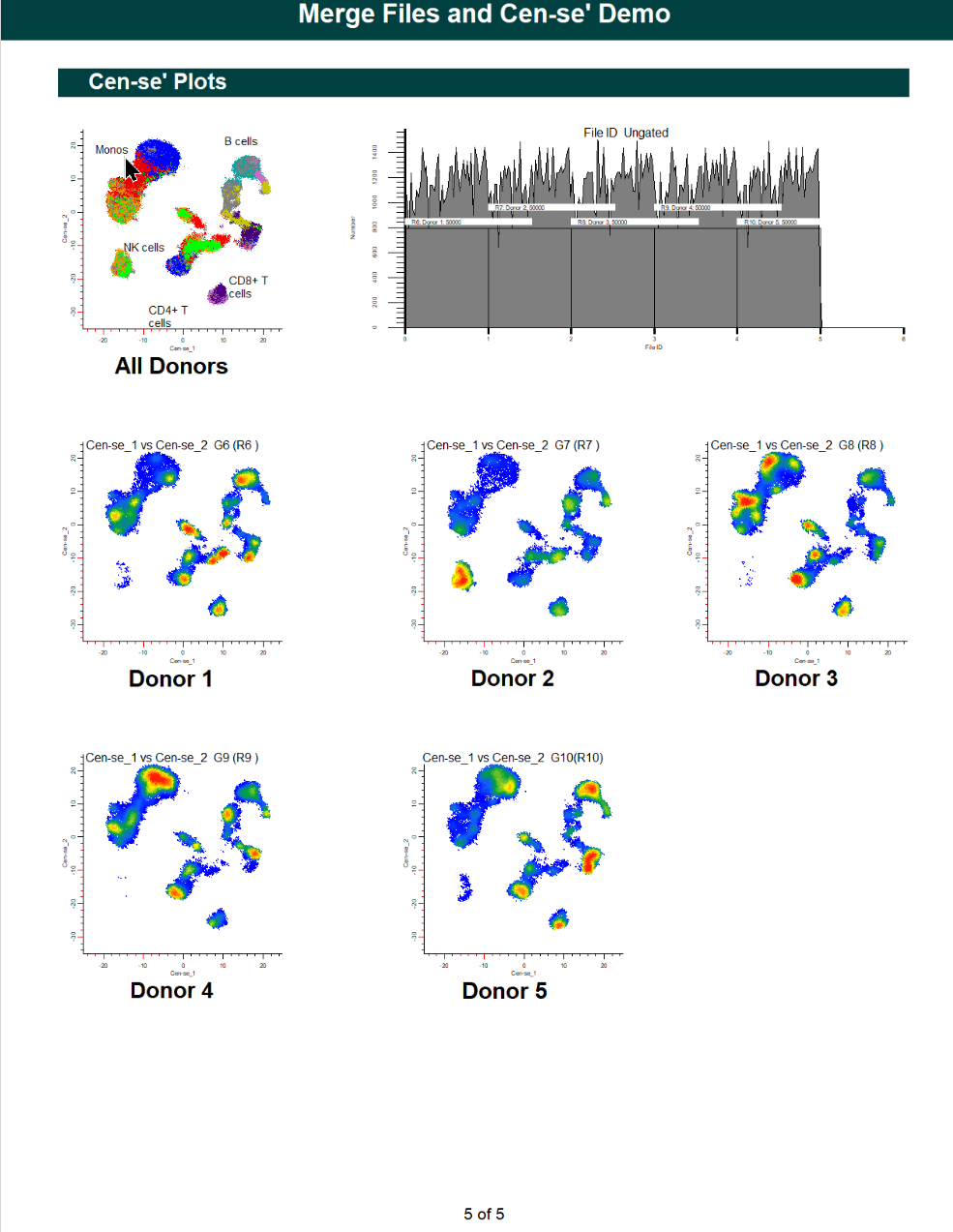
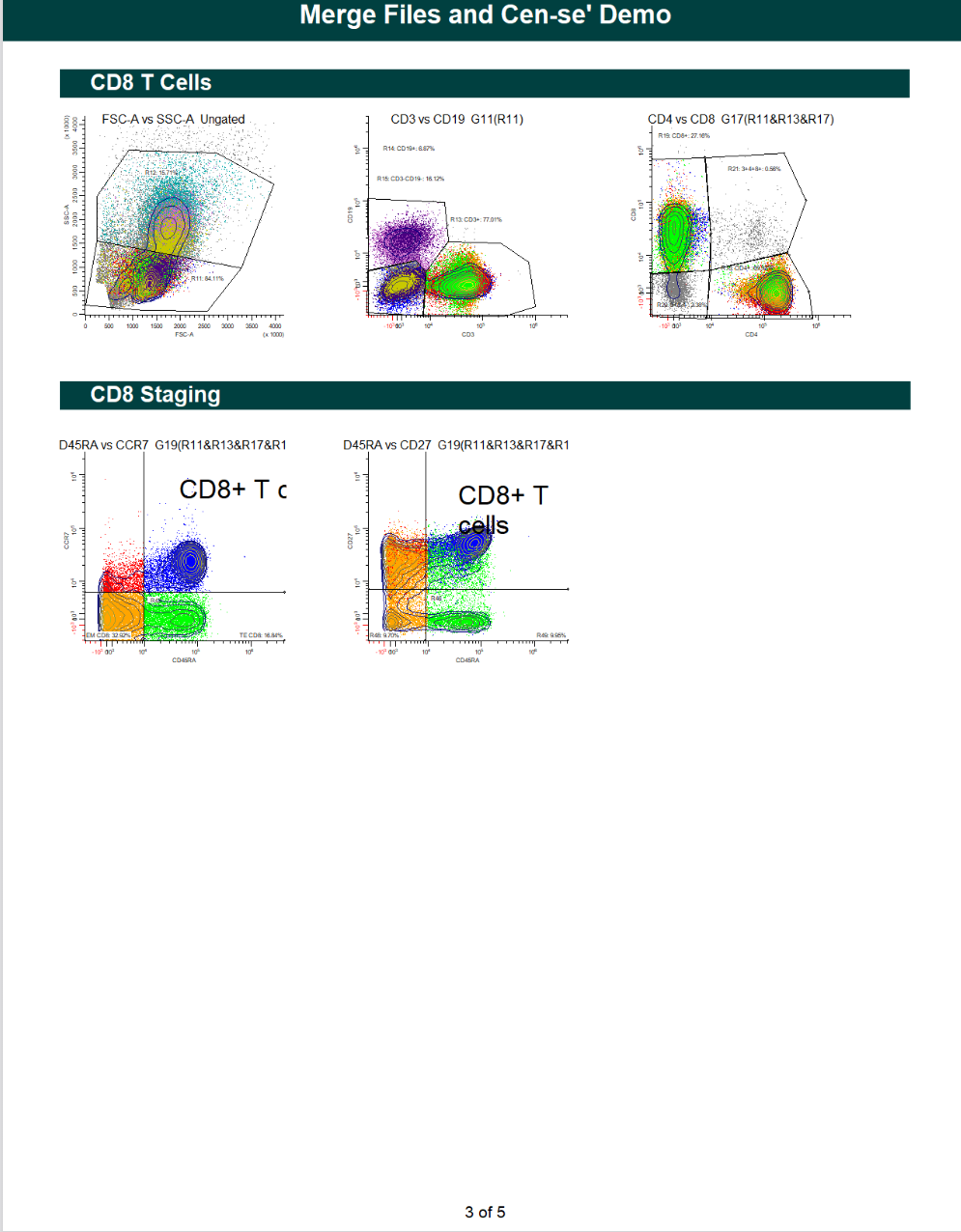
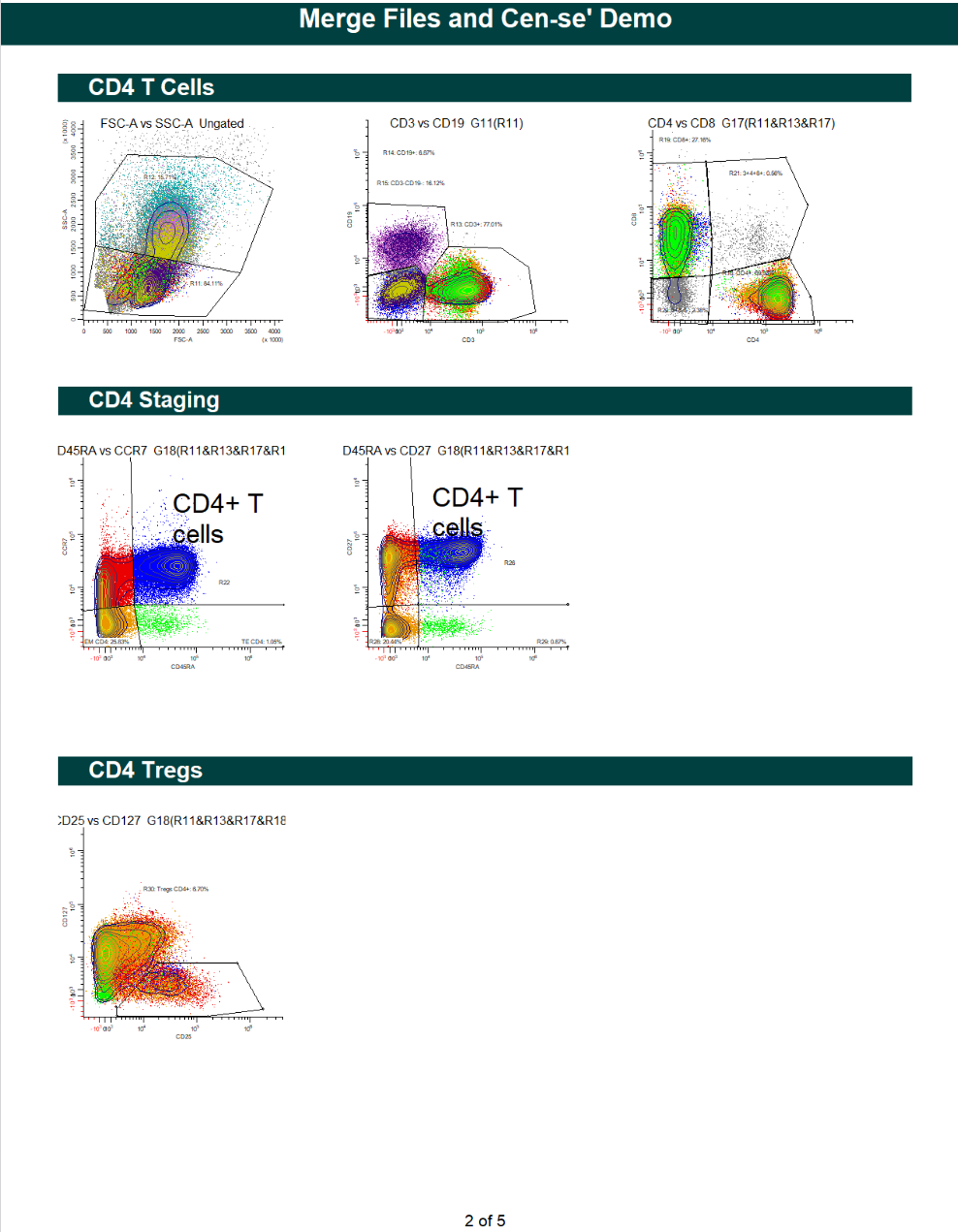
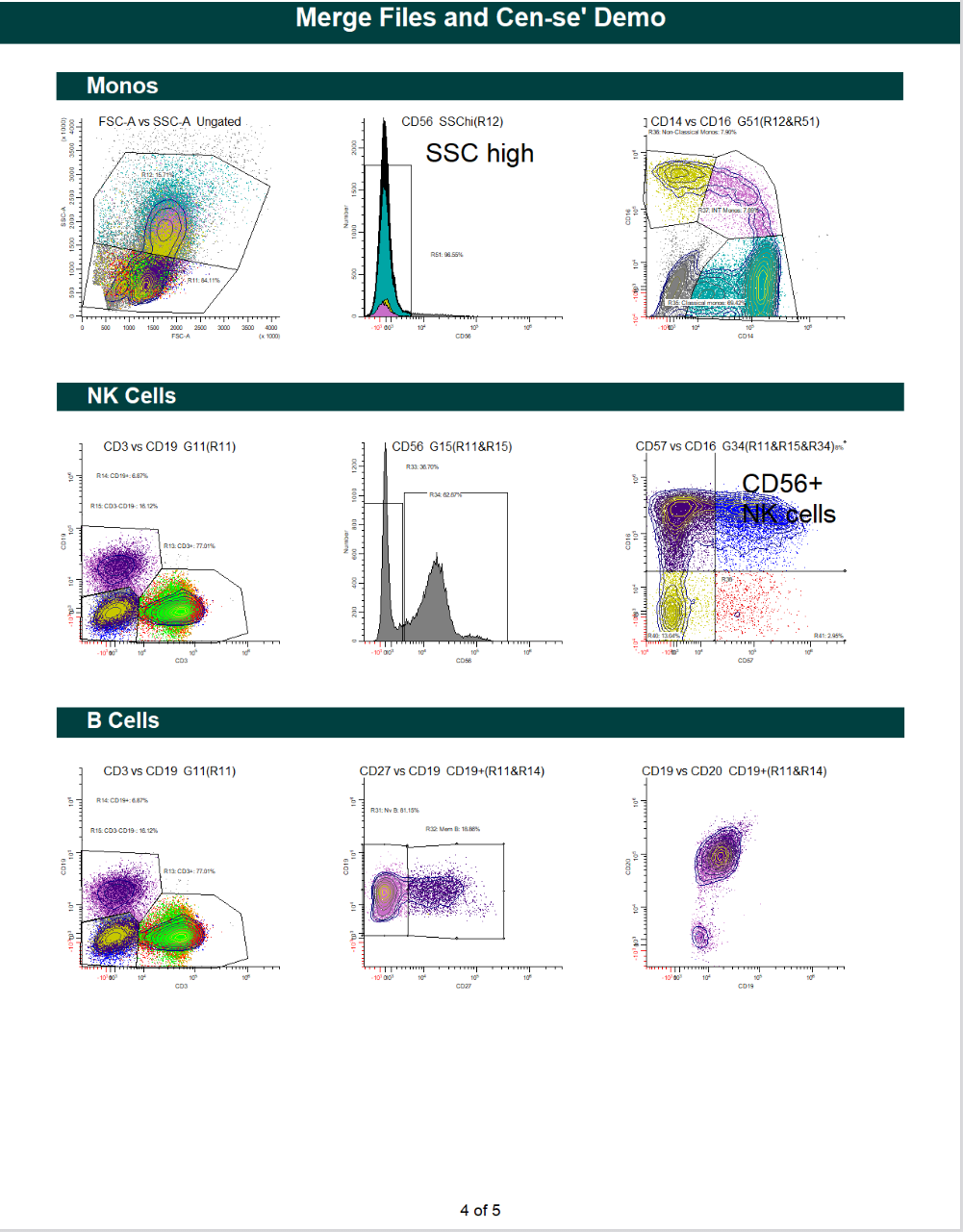
WinList Bundle shown in Merging Files and Cen-se Video
This bundle is a companion to the analysis shown in the Merging Files and Cen-se' Analysis in WinList webinar.
The video demonstrates how to merge files, and then analyzes them with Cen-se' plots.
For WinList 10+

ModFit LT Application Examples
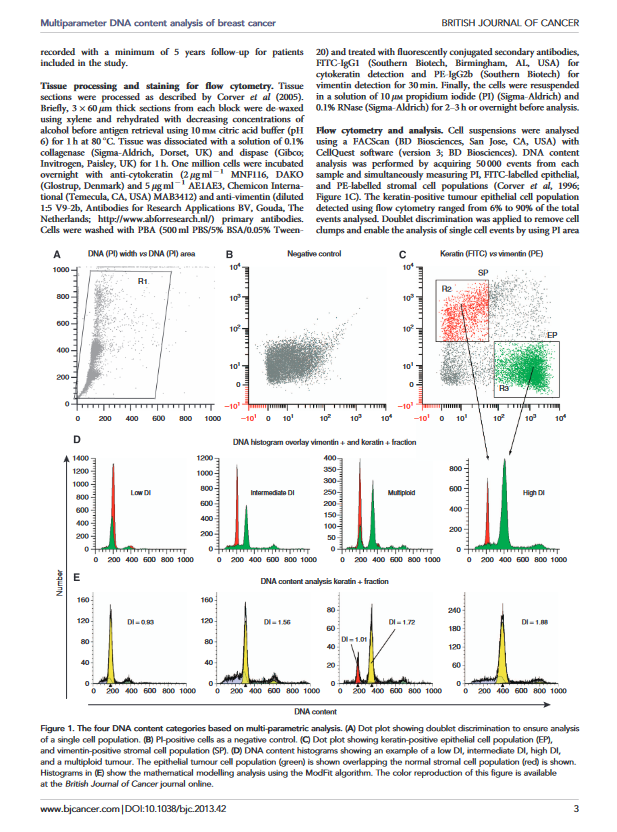
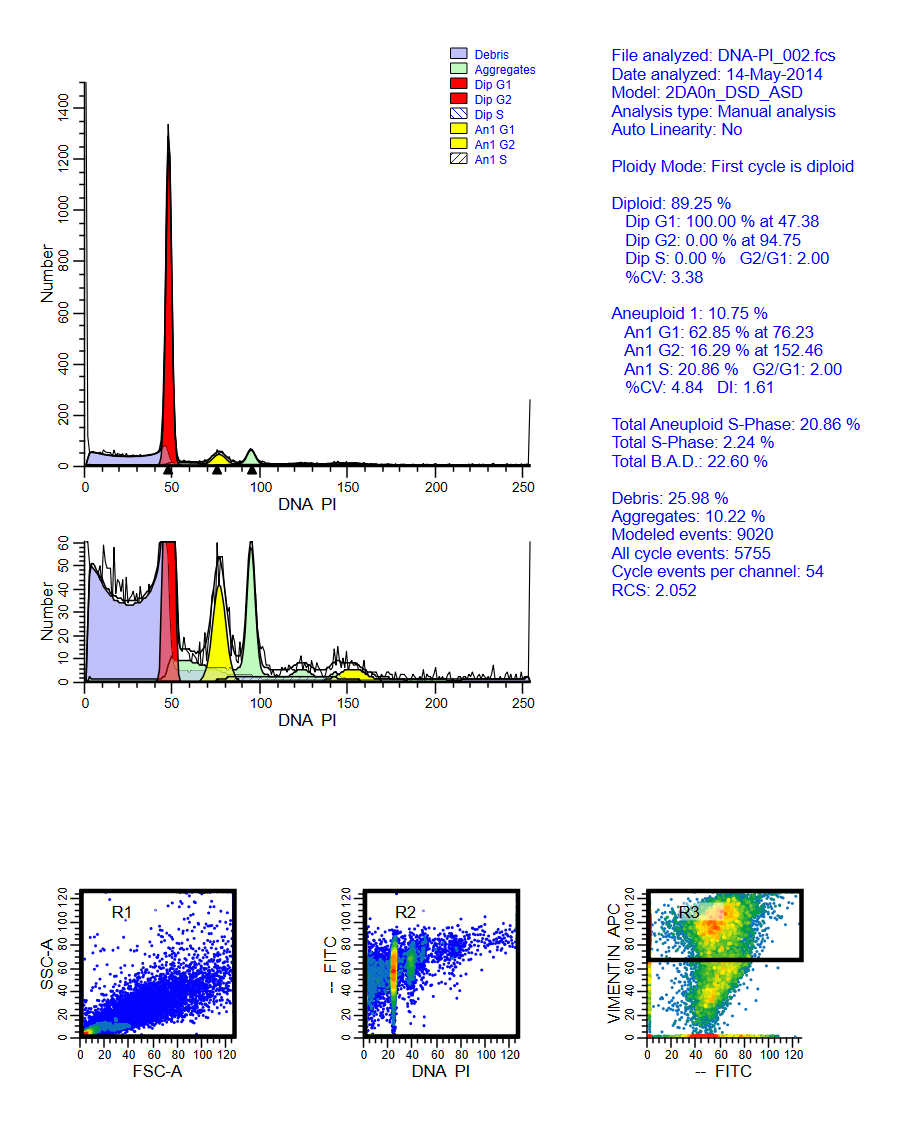
Multiparameter DNA content analysis identifies distinct groups in primary breast cancer
This study by J H S Dayal, W E Corver and others demonstrates the use of multi-parameter DNA analysis in breast cancer studies. The study was published in the British Journal of Cancer (2013), 1–8 | doi: 10.1038/bjc.2013.42
The download contains analysis of 4 samples from the study, showing the gating and ModFit analysis for each. A reprint of the paper is included.
For ModFit LT 5.0+
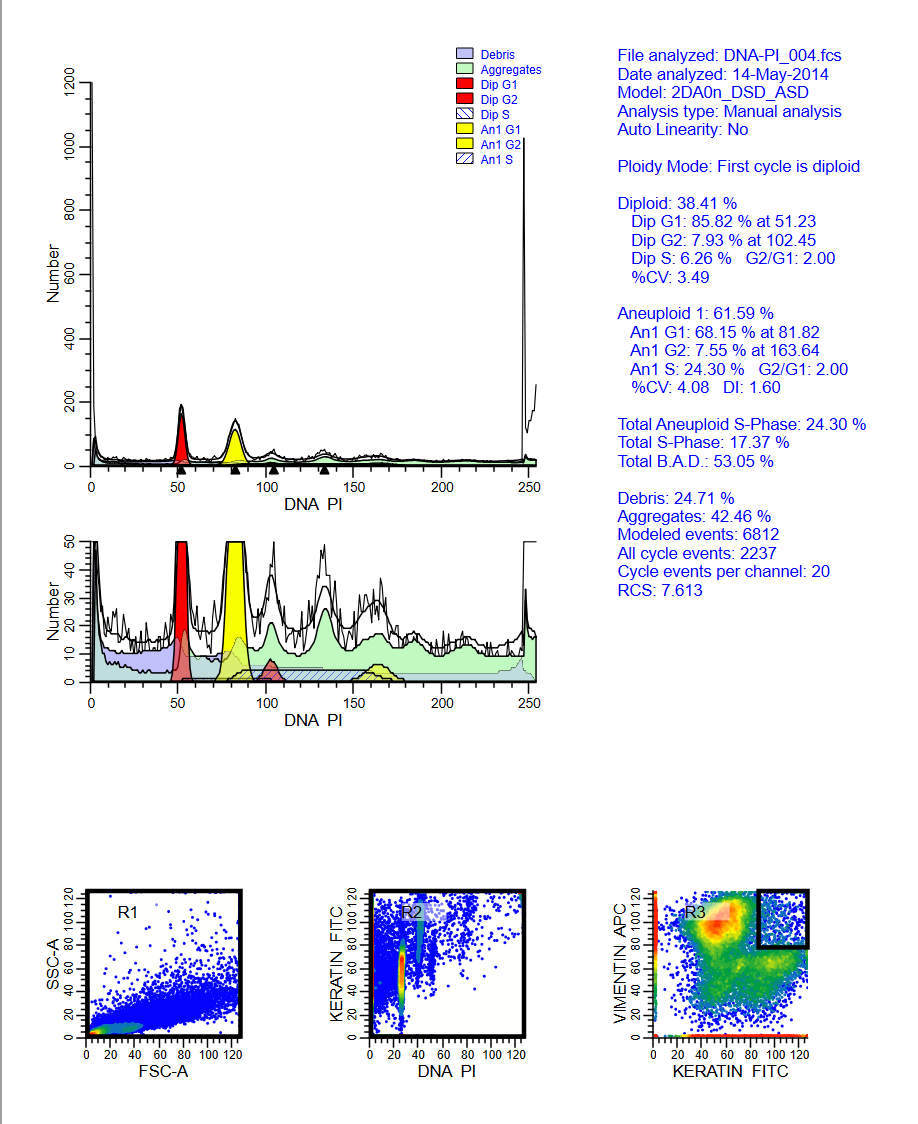
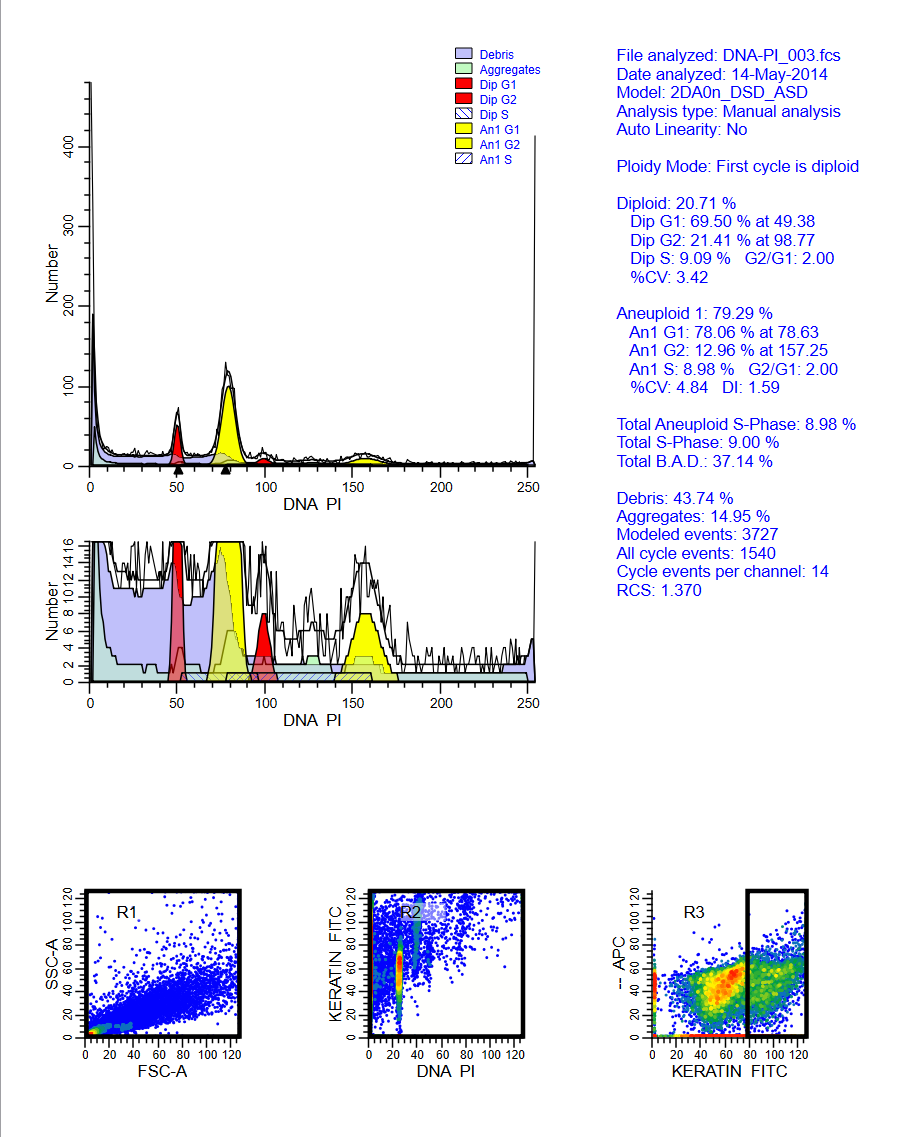
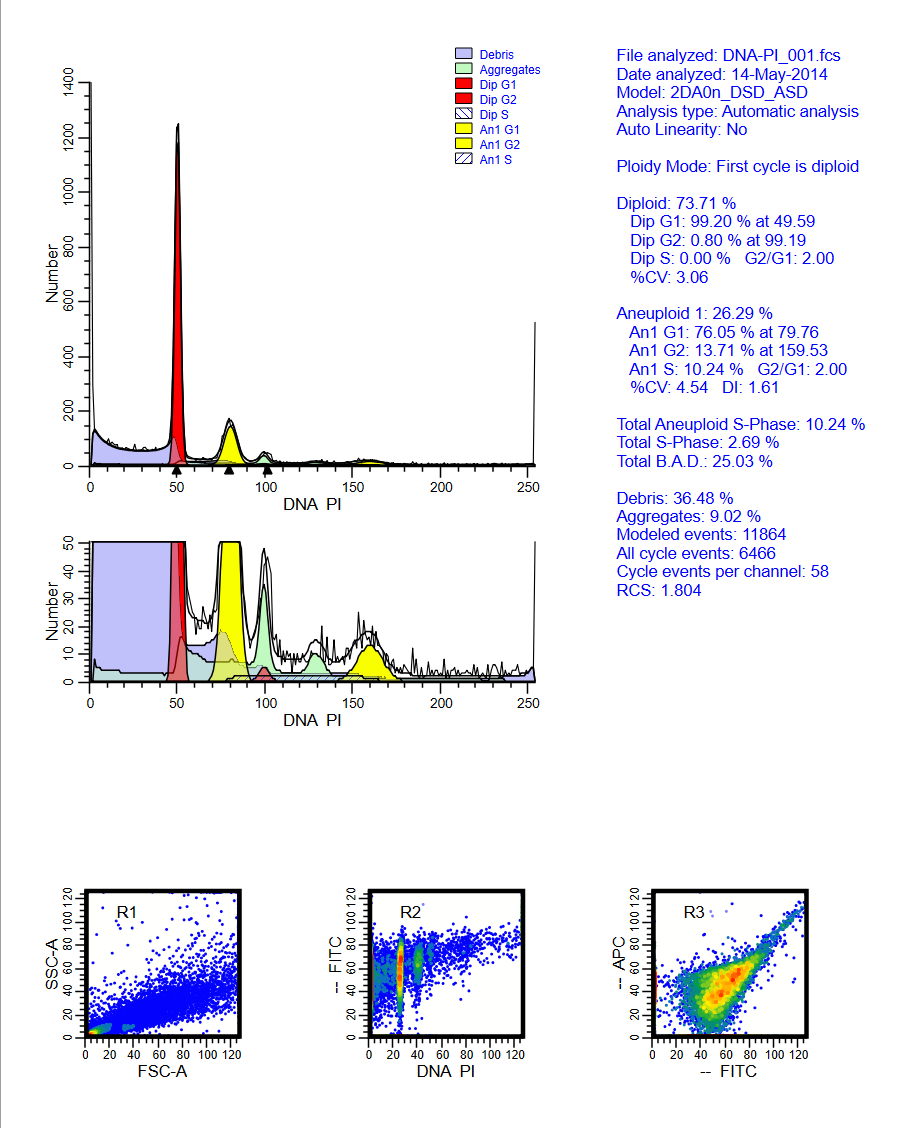
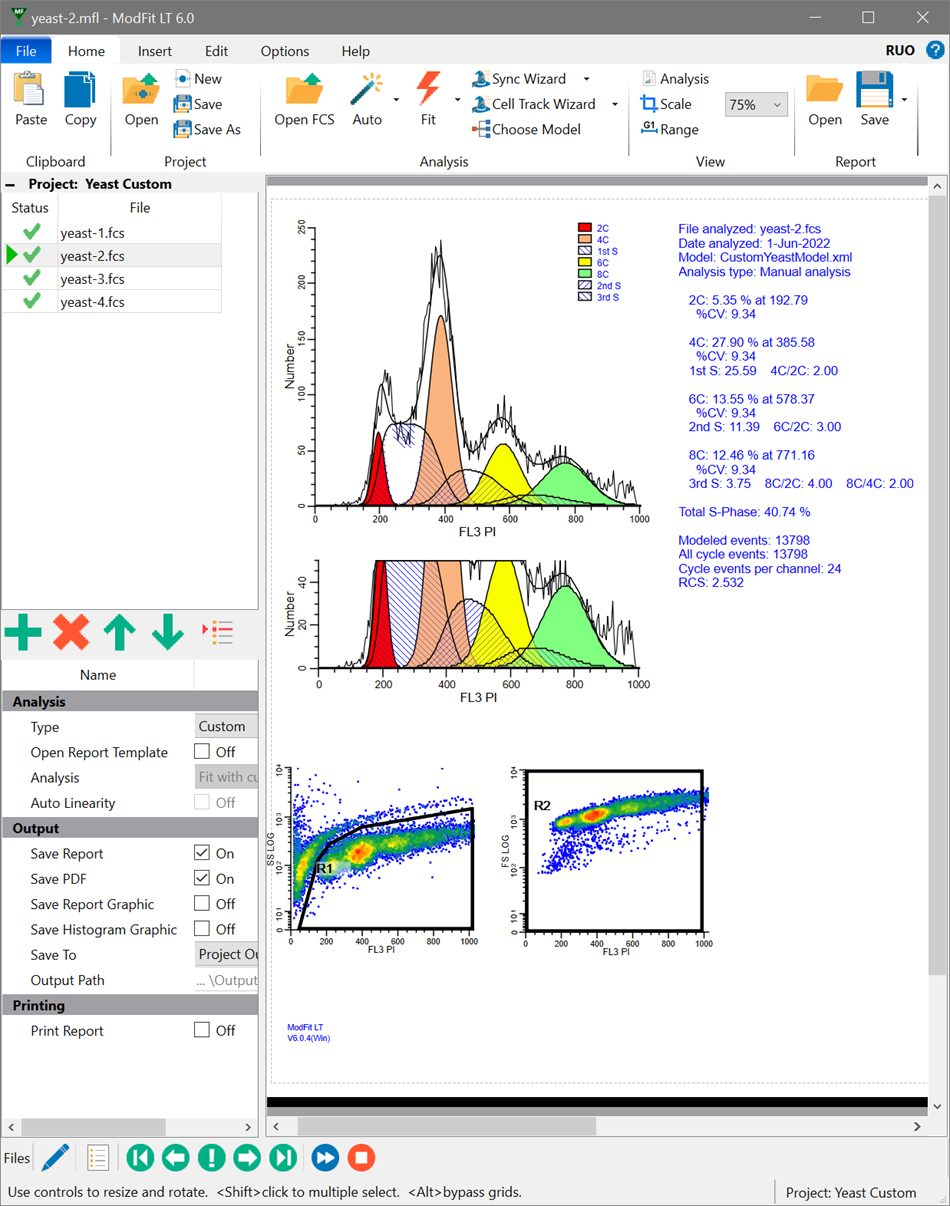
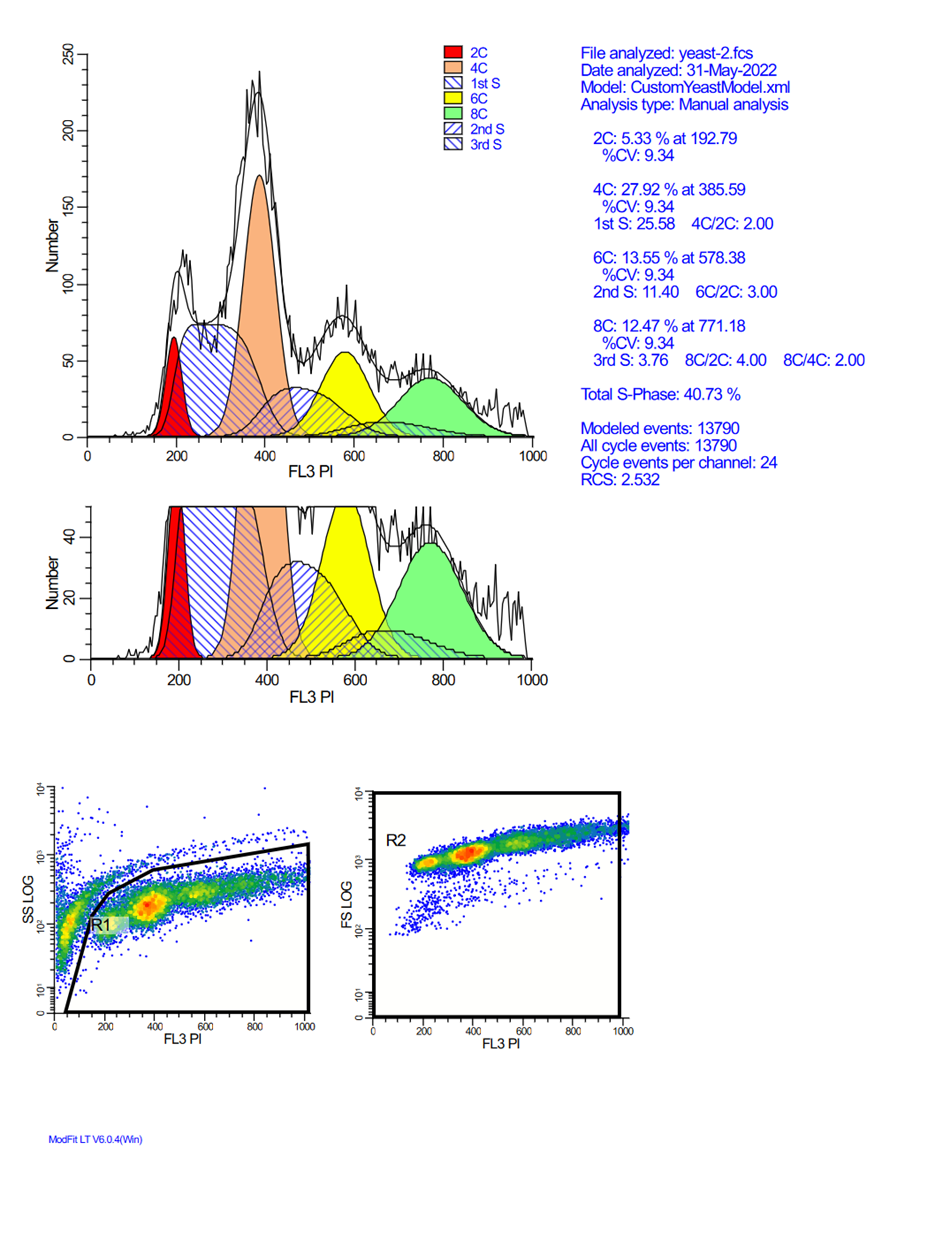
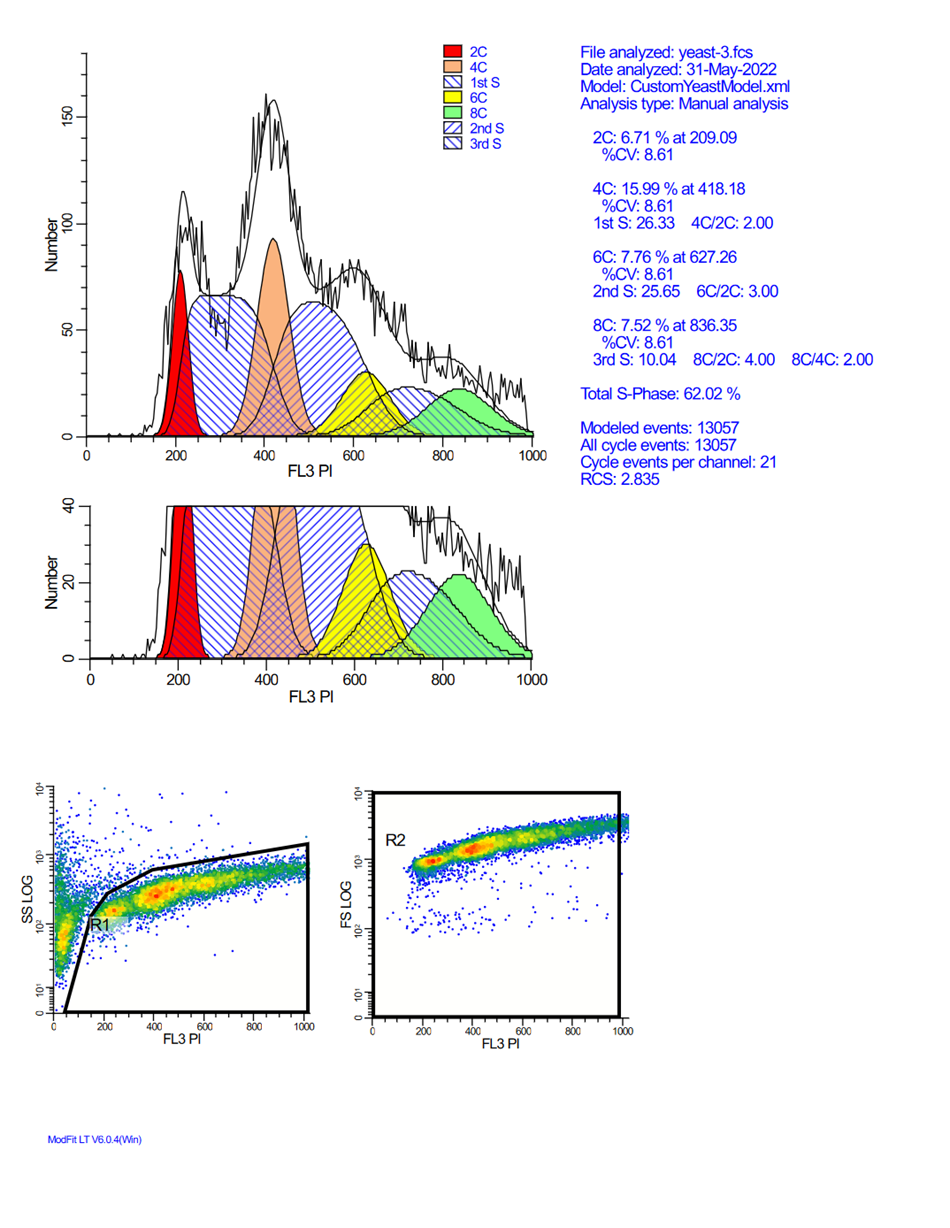
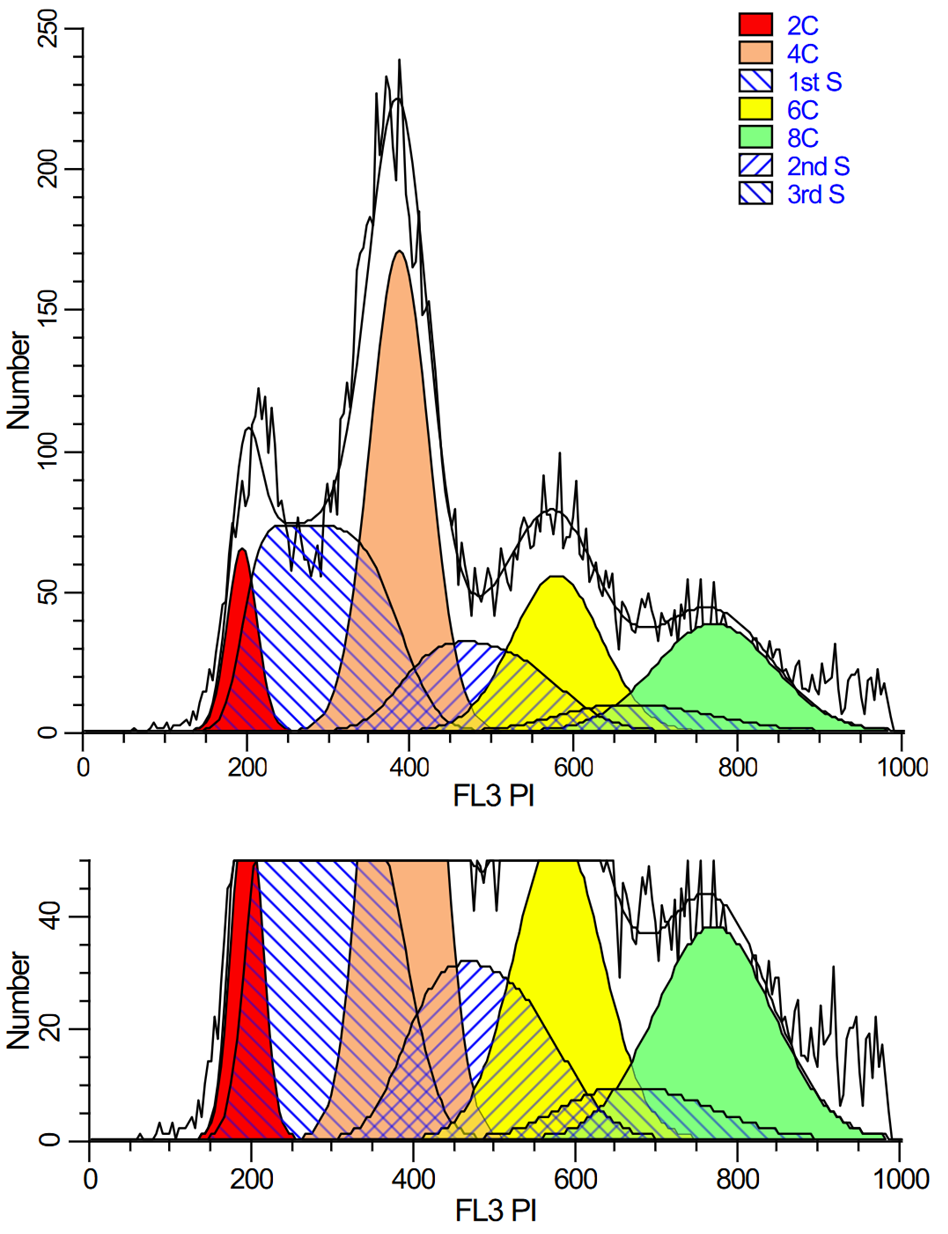
Analyzing yeast cell cycle with ModFit LT
This ModFit LT 6.0 project contains a custom model that analyzes the unique characteristics of yeast reproduction and cell cycle. The model includes haploid, 2C, 4C, 6C, and 8C populations, as well as S-Phase between each peak. This project is part of the ModFit LT 6.0 user guide tutorials.
The download contains analysis of 4 yeast samples, showing the gating and ModFit analysis for each.
For ModFit LT 6.0+